Article Detail
Population Genetics of the Short-Beaked Garfish Belone svetovidovi in Turkish Marine Waters using Mitochondrial DNA Markers
Keywords:
Belone svetovidovi
mitochondrial DNA
COI
12S rRNA
population structure
Abstract
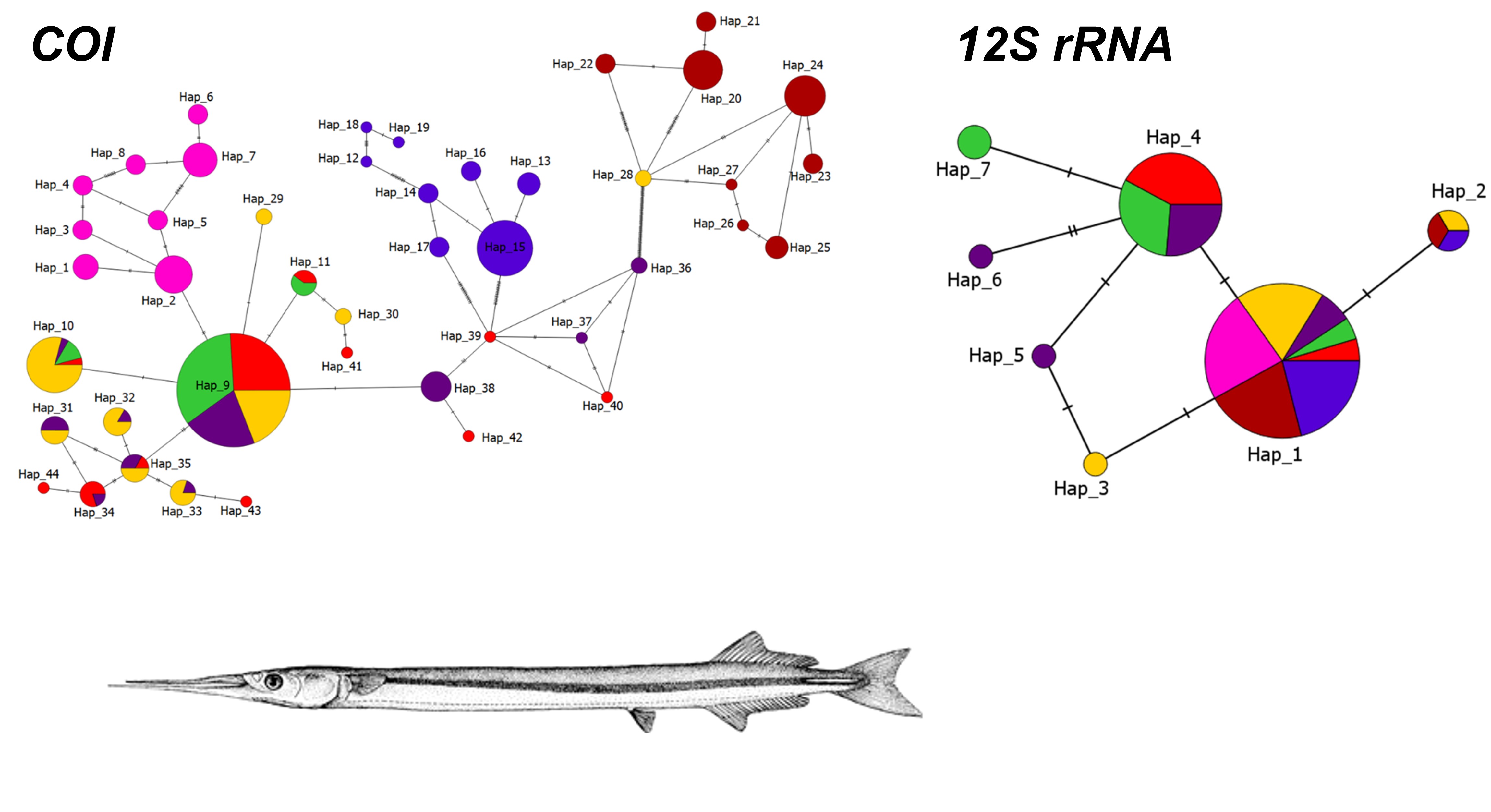
This study investigates the population genetic structure of the short-beaked garfish Belone svetovidovi across the Turkish marine coasts using two mitochondrial markers, cytochrome oxidase I and 12S ribosomal RNA. A total of 280 individuals were sampled from seven sites across the Black Sea, Marmara Sea, and Aegean Sea. Haplotype network analyses showed that COI provided greater resolution, displaying a star-like topology that points to recent population expansion, while 12S rRNA exhibited more conserved patterns, aligning with purifying selection. Tajima’s D values diverged between loci, indicating potential population structuring and demographic processes. Overall, COI captured among-basin differentiation, while 12S rRNA supported deep lineage splits. These findings highlight the complementary nature of mtDNA markers and provide baseline data for future phylogeographic and conservation studies in Belonidae species.
References
- Banford, H. M., Bermingham, E., Collette, B. B. (2004). Molecular phylogenetics and biogeography of transisthmian and amphi-Atlantic needlefishes (Belonidae: Strongylura and Tylosurus): perspectives on New World marine speciation. Molecular Phylogenetics and Evolution, 31(3), 833-851.
- Brown, K. L. (1986). Population demographic and genetic structure of plains killifish from the Kansas and Arkansas river basins in Kansas. Transactions of the American Fisheries Society, 115(4), 568-576.
- Cawthorn, D. M., Steinman, H. A., Witthuhn, R. C. (2012). Evaluation of the 16S and 12S rRNA genes as universal markers for the identification of commercial fish species in South Africa. Gene, 491(1), 40-48.
- Çetin, C., Furman, A., Kalkan, E., Bilgin, R. (2022). Mitonuclear genetic patterns of divergence in the marbled crab, Pachygrapsus marmoratus (Fabricius, 1787) along the Turkish seas. Plos One, 17(4), e0266506.
- Doğdu, S. A., Turan, C. (2021). Authentication and Traceability of Pufferfish Species Using DNA Sequencing. Pakistan Journal of Marine Sciences, 30(1), 1-11.
- Fontes, J. T., Katoh, K., Pires, R., Soares, P., Costa, F. O. (2024). Benchmarking the discrimination power of commonly used markers and amplicons in marine fish (e) DNA (meta) barcoding. ARPHA Preprints, 5, e128777.
- Fricke, R., Eschmeyer, W. N., Fong, J. D. (2025). Species by Family/Subfamily. http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp, version (04/2025).
- Froese, R., Pauly, D. (2025). FishBase. World Wide Web electronic publications. www.fishbase.org, version (06/2025).
- Galtier, N., Nabholz, B., Glémin, S., Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology, 18(22), 4541-4550.
- Gonzalez, E. G., Beerli, P., Zardoya, R. (2008). Genetic structuring and migration patterns of Atlantic bigeye tuna, Thunnus obesus (Lowe, 1839). BMC Evolutionary Biology, 8(1), 252.
- Grant, W. A. S., Bowen, B. W. (1998). Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89(5), 415-426.
- Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 41, 95-98.
- Harrison, R. G. (1989). Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends in Ecology & Evolution, 4(1), 6-11.
- Hurst, G. D., Jiggins, F. M. (2005). Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proceedings of the Royal Society B: Biological Sciences, 272(1572), 1525-1534.
- Imsiridou, A., Minos, G., Kokokiris, L., Alexandrou, M., Kyriakidou, M., Karidas, T. (2016). Genetic and phenotypic identification of Tylosurus acus imperialis in Thermaikos Gulf, North Aegean Sea. Cahiers de Biologie Marine, 57, 9-15.
- Irmak, E., Özden, U. (2023). Expansion of the flat needlefish Ablennes hians (Valenciennes, 1846) distribution in the eastern Mediterranean. Acta Adriatica, 64(1), 91-92.
- Ivanova, P., Dzhembekova, N., Atanassov, I., Rusanov, K., Raykov, V., Zlateva, I., Yankova, M., Raev, Y., Nikolov, G. (2021). Genetic diversity and morphological characterisation of three turbot (Scophthalmus maximus L., 1758) populations along the Bulgarian Black Sea coast. Nature Conservation, 43, 123-146.
- Jarosz, E., Teague, W. J., Book, J. W., Beşiktepe, Ş. T. (2013). Observed volume fluxes and mixing in the Dardanelles Strait. Journal of Geophysical Research: Oceans, 118(10), 5007-5021.
- Joseph, J., Sreedharan, S., Anoop, V. S., George, S., Antony, M. M. (2019). A preliminary investigation on the population genetic structure of Etroplus canarensis Day, 1877 of the Western Ghats, India. Asian Fisheries Science, 32, 190-195.
- Jukes, T. H., Cantor, C. R. (1969). Evolution of protein molecules. Mammalian Protein Metabolism, 3(21), 132.
- Karataş, A., Filiz, H., Erciyas-Yavuz, K., Özeren, S. C., Tok, C. V. (2021). The vertebrate biodiversity of Turkey. In Biodiversity, Conservation and Sustainability in Asia: Volume 1: Prospects and Challenges in West Asia and Caucasus (pp. 175-274). Springer.
- Kocher, T. D., Thomas, W. K., Meyer, A., Edwards, S. V., Pääbo, S., Villablanca, F. X., Wilson, A. C. (1989). Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proceedings of the National Academy of Sciences, 86(16), 6196-6200.
- Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547-1549.
- Kutlu, Y., Turan, C. (2018). An intelligent software for measurements of biological materials: BioMorph. Natural and Engineering Sciences, 3(2), 225-233.
- Lecomte, F., Grant, W. S., Dodson, J. J., Rodríguez‐Sánchez, R., Bowen, B. W. (2004). Living with uncertainty: genetic imprints of climate shifts in East Pacific anchovy (Engraulis mordax) and sardine (Sardinops sagax). Molecular Ecology, 13(8), 2169-2182.
- Leigh, J. W., Bryant, D., Nakagawa, S. (2015). POPART: full-feature software for haplotype network construction. Methods in Ecology & Evolution, 6(9), 1110–1116.
- Liu, G., Zhou, J., Zhou, D. (2012). Mitochondrial DNA reveals low population differentiation in elongate loach, Leptobotia elongata (Bleeker): implications for conservation. Environmental Biology of Fishes, 93(3), 393-402.
- Lovejoy, N. R., Collette, B. B. (2001). Phylogenetic relationships of New World needlefishes (Teleostei: Belonidae) and the biogeography of transitions between marine and freshwater habitats. Ichthyology & Herpetology, 2001(2), 324-338.
- Meriç, N., Altun, Ö. (1999). The garfish, Belone svetovidovi Collette and Parin, 1970, new to the Aegean Sea. Israel Journal of Zoology, 45, 423-426.
- Mgeleka, S. S., Silas, M. O., Mtonga, C., Rumisha, C., Viinamäki, E., Polte, P., Sköld, M., Winder, M., Gullström, M. (2023). Population genetics of the hound needlefish Tylosurus crocodilus (Belonidae) indicate high connectivity in Tanzanian coastal waters. Marine Biology Research, 19(4-5), 261-270.
- Mills, C. E., Hadwen, W. L., Hughes, J. M. (2008). Looking through glassfish: marine genetic structure in an estuarine species. Marine and Freshwater Research, 59(7), 627-637.
- Nei, M. (1987). Molecular evolutionary genetics. Columbia University Press.
- Nei, M., Tajima, F. (1981). DNA polymorphism detectable by restriction endonucleases. Genetics, 97(1), 145-163.
- Oğuz, T. (2005). Hydraulic adjustments of the Bosphorus exchange flow. Geophysical Research Letters, 32(6), L06604.
- Öztürk, D. S. (2023). Molecular and phenotypic characteristics of short-beaked garfish Belone svetovidovi Collette and Parin, 1970 in a new location, the Sea of Marmara. Marine Science and Technology Bulletin, 12(2), 156-161.
- Palsbøll, P. J., Berube, M., Allendorf, F. W. (2007). Identification of management units using population genetic data. Trends in Ecology & Evolution, 22(1), 11-16.
- Rogers, A. R., Harpending, H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 9(3), 552-569.
- Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., Sánchez-Gracia, A. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12), 3299-3302.
- Sambrook, J., Fritsch, E. F., Maniatis, T. (1989). Molecular cloning: a laboratory manual. Cold Spring Habor Laboratory.
- Schneider, S. (2000). Arlequin ver 2: A software for population genetics data analysis. University of Geneva.
- Sözer, A., Özsoy, E. (2019). Hydrodynamic Model Analysis of an Idealized Bosphorus Case with respect to Switching Steady-State Solutions. Ordu University Journal of Science and Technology, 9(1), 1-17.
- Tajima, F. (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics, 105(2), 437-460.
- Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123(3), 585-595.
- Tamura, K., Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10(3), 512-526.
- Tarula-Marin, A., Díaz-Cárdenas, B., Castro-Félix, L. P., López-Uriarte, E., Santerre, A., Aréchiga-Palomera, M. A. (2024). Genetic diversity, population structure and demographic history of the rock oyster Striostrea prismatica (Gray, 1825) within two Eastern Pacific biogeographic realms. Marine and Freshwater Research, 75(9).
- Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673-4680.
- Turan, C., Gürlek, M., Ergüden, D., Yağlıoğlu, D., Uyan, A., Reyhaniye, A. N., Özbalcılar, B., Öztürk, B., Erdoğan, Z., Ivanova, P., Soldo, A. (2015). Population genetic analysis of Atlantic bonito Sarda sarda (Bloch, 1793) using sequence analysis of mtDNA D-loop region. Fresenius Environmental Bulletin, 24(10), 3148-3154.
- Turan, C., Ivanova, P., Soldo, A. (2016). Population structuring and migration pathway of Atlantic bonito Sarda sarda. Natural and Engineering Sciences, 1(3), 56-65.
- Turan, C., Yağlıoğlu, D., Doğdu, S. A., Ergüden, D., Ivanova, P. P., Raykov, V. S. (2023a). Existence of Belone svetovidovi Collette & Parin, 1970 (Family: Belonidae) in the Marmara Sea and Black Sea Coasts of Türkiye. Natural and Engineering Sciences, 8(2), 72-81.
- Turan, C., Uyan, A., Atalay, M. A., Doğdu, S. A., Ayas, D., Ergüden, D., Turan, F., Gökoğlu, M., Gürlek, M. (2023b). Genetic differentiation of round sardinella Sardinella aurita (Clupeidae) populations from the Northeastern Mediterranean. Journal of Ichthyology, 63(5), 962-968.
- Turan, C., Ivanova, P., Doğdu, S., Ergüden, D., Raykov, V., Yankova, M., Yağlioğlu, D., Ergenler, A., Zlateva, I., Dzhembekova, N. (2024). Population genetic analysis of garfish Belone belone from the Bulgarian and Turkish coasts of the Black Sea. Tethys Environmental Science, 1(3), 152-163.
- Turan, C., Ivanova, P. P., Doğdu, S. A., Ergüden, D., Raykov, V. S., Yankova, M., Yağlıoğlu, D., Ergenler, A. (2025). Systematic identification of needlefish (Belonidae) species using molecular genetic and morphological markers in the Mediterranean and Black Seas. PloS One, 20(2), e0315401.
- Ward, R. D. (2000). Genetics in fisheries management. Hydrobiologia, 420(1), 191-201.
- Wei, H., Geng, L., Shang, X., Li, L., Ma, B., Zhang, Y., Li, W., Xu, W. (2023). Comparison genetic diversity and population structure of four Pseudaspius leptocephalus populations in Heilongjiang River Basin based on mitochondrial COI gene. Frontiers in Marine Science, 10, 1158845.
- Wilson, A. J., Gislason, D., Skúlason, S., Snorrason, S. S., Adams, C. E., Alexander, G., Danzmann, R. G., Ferguson, M. M. (2004). Population genetic structure of Arctic charr, Salvelinus alpinus from northwest Europe on large and small spatial scales. Molecular Ecology, 13(5), 1129-1142.
- Xu, H., Huang, L., Chen, T., Wang, C., Wu, Z., Cheng, Y., Su, Q., Kang, B., Yan, Y., Zhang, X. (2024). Phylogeography and population structure of Lagocephalus spadiceus (Richardson, 1845)(Tetraodontiformes, Tetraodontidae) in the South China Sea. Ecology and Evolution, 14(4), e11320.
- Xu, H., Zhang, Y., Xu, D., Lou, B., Guo, Y., Sun, X., Guo, B. (2014). Genetic population structure of miiuy croaker (Miichthys miiuy) in the Yellow and East China Seas base on mitochondrial COI sequences. Biochemical Systematics and Ecology, 54, 240-246.
- Yankova, M., Raykov, V., Ivanova, P., Dzhembekova, N., Turan, C., Raev, Y. (2023). Morphological and genetic characteristics of garfish Belone belone (L., 1760)(Belonidae, Teleostei) population from the southern Bulgarian Black Sea coast. Nature Conservation, 54, 1-12.
- Ye, Y., Fu, Z., Tian, Y., Li, J., Guo, B., Lv, Z., Wu, C. (2018). Pelagic larval dispersal habits influence the population genetic structure of clam Gomphina aequilatera in China. Genes & Genomics, 40(11), 1213-1223.
35
16381
95515
20days
50days
10days
